Due to significant improvements in genomic sequencing technology, it is now possible to sequence a full human genome for less than $1000 and receive genetic data on several molecular levels of the cell (genomic, transcriptome, protein expression, copy-number variations, and so on). However, in order to turn this genomic Big Data into useful insights for precision medicine, a systematic integrated study of a range of biological datasets is required. ACCELOM develops bespoke, reproducible multi-omics software pipelines that combine numerous molecular datasets from a host to investigate association patterns between molecular levels using unique, peer-reviewed statistical machine learning algorithms.
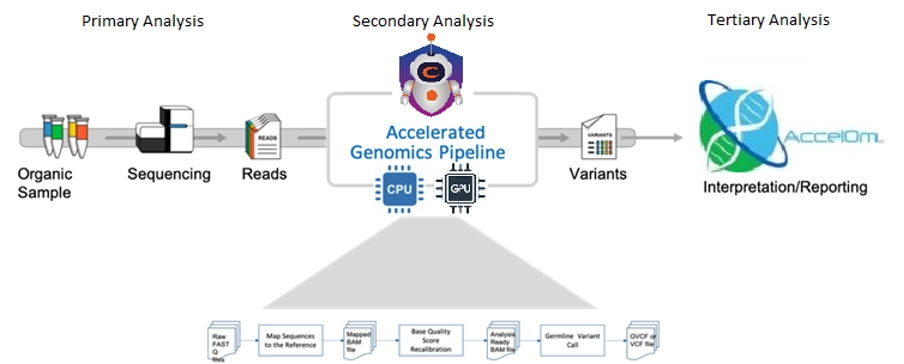
ACCELOM pipelines rely on the well-established GATK pipeline for performing secondary analysis, which transforms raw sequenced reads into analysis-ready variants. It is well known that the GATK pipeline is extremely computationally intensive[1] especially for large datasets (ex. Whole Genome Sequencing). To address this bottleneck, several hardware acceleration solutions have emerged to scale secondary analysis[2] [3]. While these solutions often improve time-to-insight by several orders of magnitude, they are closed and proprietary on both the software front (as accelerated pipelines not freely available) and hardware front (as they are tied to a specific vendor hardware).
Through SYCLOPS, ACCELOM intends to collaborate with various partners in the development and validation of a SYCL-based library (SYCL-GAL) that will accelerate key stages of the GATK secondary analysis pipeline. Unlike contemporary solutions, SYCL-GAL library will be open source in nature to foster the adoption of open standards in genomics. Further, by relying on SYCL for cross-architecture parallelism, SYCL-GAL will also enable cross-platform acceleration of genomic data analysis, making it possible to deploy the same pipeline on CPUs, GPUs, and accelerators from several hardware vendors.

 This Horizon Europe Project "SYCLOPS" has received funding from the European Union HE Research and
Innovation programme under grant agreement No 101092877. Codeplay Software, as a UK participant
in this project is supported by UK Research and Innovation Scheme grant numbers 10048920.
This Horizon Europe Project "SYCLOPS" has received funding from the European Union HE Research and
Innovation programme under grant agreement No 101092877. Codeplay Software, as a UK participant
in this project is supported by UK Research and Innovation Scheme grant numbers 10048920.

